Learning by Doing
 The goal of post-secondary science education is to
train future scientists. One problem with science education is the standard lecture/laboratory format. In
lecture the professor speaks, and the student passively listens. In the worst case, lectures are no better
for learning than television: a totally passive, non-interactive experience. Meanwhile, laboratories are
intended to afford students with an interactive, experimental experience, but in reality these are usually
rigidly structured by the laboratory outline where the intended outcome is known and the procedure is
inflexible. These are not experimental experiences. If laboratories were to be truly experimental,
question- and hypothesis-driven experiences, then students would need access to modern laboratory
equipment and instrumentation, a vast array of expensive laboratory reagents, and a wealth of instructor
time that the university cannot possibly afford. Active learning can be expensive, yet the value of active
versus passive learning has become increasingly clear (Reid, 1994).
The goal of post-secondary science education is to
train future scientists. One problem with science education is the standard lecture/laboratory format. In
lecture the professor speaks, and the student passively listens. In the worst case, lectures are no better
for learning than television: a totally passive, non-interactive experience. Meanwhile, laboratories are
intended to afford students with an interactive, experimental experience, but in reality these are usually
rigidly structured by the laboratory outline where the intended outcome is known and the procedure is
inflexible. These are not experimental experiences. If laboratories were to be truly experimental,
question- and hypothesis-driven experiences, then students would need access to modern laboratory
equipment and instrumentation, a vast array of expensive laboratory reagents, and a wealth of instructor
time that the university cannot possibly afford. Active learning can be expensive, yet the value of active
versus passive learning has become increasingly clear (Reid, 1994).
The Virtual Cell
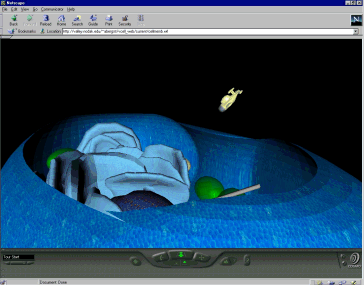 The Virtual Cell is a virtual, multi-user space where students
"fly" around and practice being cell biologists in a role-based, goal-oriented environment. Working
individually, or with others, students learn fundamental concepts of cell biology and strategies for
deductive problem solving through their experiences in the exploratory environment. This pedagogical
approach gives students an authentic experience that includes elements of practical, experimental
design and decision making, while introducing them to discipline content. By practical applications of the
scientific method, students learn how to think, act, and react as cell biologists (Slator and Chaput, 1996).
The Virtual Cell is a virtual, multi-user space where students
"fly" around and practice being cell biologists in a role-based, goal-oriented environment. Working
individually, or with others, students learn fundamental concepts of cell biology and strategies for
deductive problem solving through their experiences in the exploratory environment. This pedagogical
approach gives students an authentic experience that includes elements of practical, experimental
design and decision making, while introducing them to discipline content. By practical applications of the
scientific method, students learn how to think, act, and react as cell biologists (Slator and Chaput, 1996).
The Virtual Cell Experience: The Laboratory
The Cell
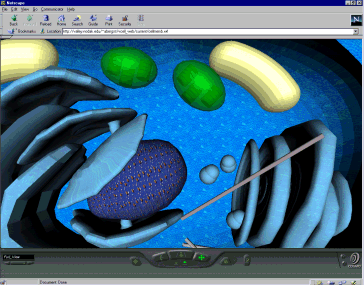 The virtual cell contains 3D representations of all the
components and organelles of a cell (nucleus, mitochondria, chloroplasts, etc). The user "flies" among
these organelles and uses virtual instruments to conduct experiments. All navigation is learner directed;
there is no predetermined exploratory path. This feature empowers the student to direct their own
learning. The student is also able to travel into linked VRML worlds that represent the interior of each of
the cellular organelles. Further experimentation inside each organelle allows the student to learn about
their specific functions (Wu, 1998).
The virtual cell contains 3D representations of all the
components and organelles of a cell (nucleus, mitochondria, chloroplasts, etc). The user "flies" among
these organelles and uses virtual instruments to conduct experiments. All navigation is learner directed;
there is no predetermined exploratory path. This feature empowers the student to direct their own
learning. The student is also able to travel into linked VRML worlds that represent the interior of each of
the cellular organelles. Further experimentation inside each organelle allows the student to learn about
their specific functions (Wu, 1998).
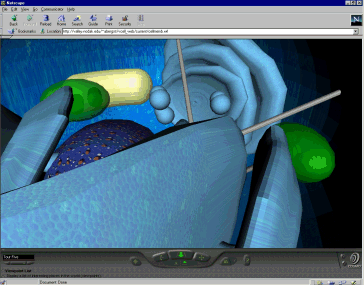 More advanced levels of the Virtual Cell include the
introduction of cellular perturbations and the investigation of the functions of various cellular structures or
processes. In the second level, a simulation will change the cell by either introducing a mutation or
adding an inhibitor that disrupts a cellular process. An alarm will sound, some cellular process will
malfunction, and the learner will be given the goal of diagnosing the problem. Using the same tools as in
the previous level, the learner will navigate through the cell, make observations, and perform
measurements and experiments. The learner will attempt to identify the affected area, the perturbed
process, and the nature of the mutation or inhibitor that is causing the problem. As a result, the user will
learn details of cellular processes and functions and become familiar with the importance of various
mutations and inhibitors for cell biology experimentation.
More advanced levels of the Virtual Cell include the
introduction of cellular perturbations and the investigation of the functions of various cellular structures or
processes. In the second level, a simulation will change the cell by either introducing a mutation or
adding an inhibitor that disrupts a cellular process. An alarm will sound, some cellular process will
malfunction, and the learner will be given the goal of diagnosing the problem. Using the same tools as in
the previous level, the learner will navigate through the cell, make observations, and perform
measurements and experiments. The learner will attempt to identify the affected area, the perturbed
process, and the nature of the mutation or inhibitor that is causing the problem. As a result, the user will
learn details of cellular processes and functions and become familiar with the importance of various
mutations and inhibitors for cell biology experimentation.
Simulations
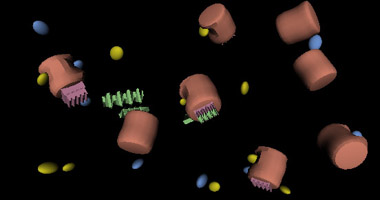
Multiuser Aspects
Summary
References
- Curtis, Pavel (1992). Mudding: Social Phenomena in Text-Based Virtual Realities. Proceedings of
the conference on Directions and Implications of Advanced Computing (sponsored by Computer
Professionals for Social Responsibility)
- Reid , T Alex (1994) Perspectives on computers in education: the promise, the pain, the prospect.
Active Learning. 1(1), Dec. CTI Support Service. Oxford, UK
- Slator, Brian M. and Harold "Cliff" Chaput (1996) Learning by Learning Roles: a virtual role-playing
environment for tutoring. Proceedings of the Third International Conference on Intelligent Tutoring
Systems (ITS'96). Montreal: Springer-Verlag, June 12-14, pp. 668-676. (Lecture Notes in Computer
Science, edited by C. Frasson, G. Gauthier, A. Lesgold)
- Wu, Yihe (1998) Simulation of the Biological Process of Steroid Hormones with VRML. M.S. Thesis. North Dakota State University, Fargo, ND
Development of the Virtual Cell is funded by the National Science Foundation under grants DUE-9752548. For further information on virtual worlds software development at North Dakota State University, visit the NDSU WWWIC web site. The authors acknowledge the large team of dedicated undergraduate and graduate students in the computer and biological sciences who have made this project so successful. © 1998 World Wide Web Instructional Committee.